Molecular phylogenetics – uncovering the history of evolution using signals in organisms’ genetic sequences – is a powerful science, the latest expression of the human desire to understand our common origins. But for all its achievements, I’d always felt something was, well lacking from my science. This week we’ve been experimenting with the MinION for the first time, and now I know what that is: immediacy. I’ll return to this theme later to explain how exciting this realisation is, but first we better ask:
Molecular phylogenetics – uncovering the history of evolution using signals in organisms’ genetic sequences – is a powerful science, the latest expression of the human desire to understand our common origins. But for all its achievements, I’d always felt something was, well lacking from my science. This week we’ve been experimenting with the MinION for the first time, and now I know what that is: immediacy. I’ll return to this theme later to explain how exciting this realisation is, but first we better ask:
What is a MinION?
NGS nerds know about this already, of course, but for the rest of us: The MinION (minh-aye-on) is a USB-connected device marketed by a UK company, Oxford Nanopore, as ‘a portable real-time biological analyser’. Yes – that does sound a lot like a tricorder, and for good reason: just like the fictional device, it promises to make the instant identification of biological samples a reality. It does this using a radically different new way to ‘read’ DNA sequences from a liquid into a letters (‘A, C, G, T‘) on a computer screen. Essentially, individual strands of DNA are pulled through a hole (a ‘nanopore’) in an artificial membrane, like the membranes that surround every living cell. When an electric field is applied to the membrane, the individual DNA letters (actually, ‘hexamers’ – 6-letter chunks) passing through the nanopore can be directly detected as fluctuations in the field, much like a magnetic C-60 cassette tape is read by a magnetic tape head. The really important thing is that, whereas other existing DNA-reading (‘sequencing’) machines take days-to-weeks to read a sample, the MinION produces output in minutes or even seconds. It’s also (as you can see in the picture, above) a small, wait tiny device.
Real-time
This combination of fast results and small size makes the tricorder dream possible, but this is more than just a gadget. Really, having access to biological sequences at our fingertips will completely change biology and society in many ways, some of which Yaniv Erlich explored in a recent paper. In particular, molecular evolutionary biologists will soon be able to interact with their subject matter in ways that other scientists have long taken for granted. What I mean by this is that in many other empirical disciplines, researchers (and schoolkids!) are able to directly observe, or easily measure, the phenomena they study. Paleontologists dig bones. Seismologists feel earthquakes. Zoologists track lions. And so on. But until now, the world of genomic data hasn’t been directly observable. In fact reading DNA sequences is a slow, expensive pain in the arse. And in such a heavily empirical subject, I can’t help but feel this is a hindrance – we are burdened with a galaxy of baroque models to explain variations in the small number of observations we’ve made, and I’ve got a hunch more data would actually consign many of them to the dustbin.
Imagine formulating, testing, and validating or discarding genetic hypotheses as seamlessly as an ecologist might survey a new forest…

Actual use
That’s the spiel, anyway. This week we actually used the MinION for the first time to sequence DNA (I’d been running some other technical tests for a couple of months, but this was our first attempt with real samples) and since it seems lots more colleagues want to ask about this device than have had a chance to use it, I thought I’d share our experiences. I say ‘we’ – work took place thanks to a Pilot Study Fund grant at the Royal Botanic Gardens, Kew, in collaboration with Dr. Alex Papadopulos (and really useful input from Drs. Andrew Helmstetter, Pepijn Kooij and Bryn Dentinger). It’s worth pointing out that although there’s a lot of hype about the device, it is still technically in a prototype/public-beta type phase, so things are changing all the time in terms of performance, etc.
 First up, the size. The pictures don’t really do it justice… compared with existing machines (fridge-sized, often) the MinION isn’t small, it’s tiny. The thing itself, plus box and cable, would easily fit into a side pocket on any laptop bag. So the ‘portable’ bit is certainly true, as far as the platform itself is concerned. But there’s a catch – to prepare a biological sample for sequencing on the MinION, you first have to go through a fairly complicated set of lab steps. Known collectively as ‘library preparation’ (a library here meaning a test tube containing a set of DNA molecules that have been specially treated to prepare them to be sequenced), the existing lab protocol took us several hours the first time. Partly that was due to the sheer number of curious onlookers, but partly because some of the requisite steps (bead cleanups etc) just need time and concentration to get right. None of the steps is particularly complicated (I’m crap at labwork and just about followed along) but there’s quite a few, and you have to follow the steps in the protocol carefully.
First up, the size. The pictures don’t really do it justice… compared with existing machines (fridge-sized, often) the MinION isn’t small, it’s tiny. The thing itself, plus box and cable, would easily fit into a side pocket on any laptop bag. So the ‘portable’ bit is certainly true, as far as the platform itself is concerned. But there’s a catch – to prepare a biological sample for sequencing on the MinION, you first have to go through a fairly complicated set of lab steps. Known collectively as ‘library preparation’ (a library here meaning a test tube containing a set of DNA molecules that have been specially treated to prepare them to be sequenced), the existing lab protocol took us several hours the first time. Partly that was due to the sheer number of curious onlookers, but partly because some of the requisite steps (bead cleanups etc) just need time and concentration to get right. None of the steps is particularly complicated (I’m crap at labwork and just about followed along) but there’s quite a few, and you have to follow the steps in the protocol carefully.
Performance
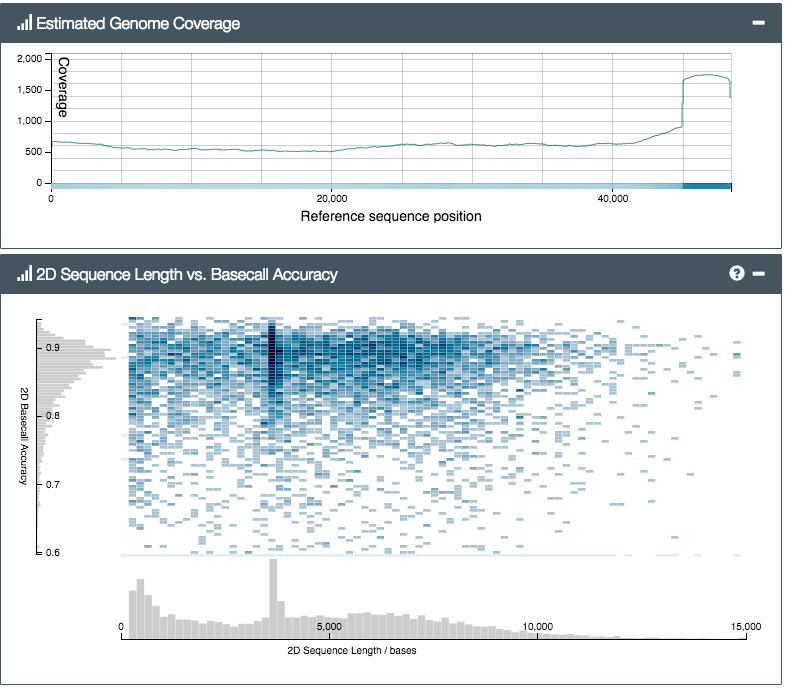
So how did the MinION do? We prepared two libraries; one from a control sample of bacteriophage lambda (a viral genome, used to check the lab steps are working) and another from Silene latifolia, a small flowering plant and one of Alex’s faves. The results were exciting. In fact they were nerve-wracking – initially we mixed up the two samples by mistake (incredible – what pros…) and were really worried when, after a few hours’ sequencing, not a single DNA read matching the lambda genome had appeared. After a lot of worrying, and restarting various analyses, including the MinION itself, we eventually realised our mistake, reloaded the MinION with the other sample and – hey presto! – lambda reads started to pour out of the software. In the end, we were able to get a 500x coverage really quickly (see reads mapped to the reference genome, below):
You can see from the top plot that we got good even coverage across the genome, while the bottom plot shows an even (ish) read length distribution, peaking around 4kb. We’d tried for a target size of 6kb (shearing using g-TUBEs), so it seems the actual output read size distribution is lower than the shearing target – you’d need to aim higher to compensate. Still, we got plenty of reads longer than 20, or even 30,000 base-pairs (bp) – from a 47kbp genome this is great, and much much much longer than typical Illumina paired-end reads of ~hundreds of bp.
Later on the next day, we decided we’d done enough to be sure the library preparation and sequencing were working well, so we switched from the lambda control to our experimental (S.latifolia) sample. Again, we got a good steady stream of reads, and some were really long, over 65,000bp. Crucially, we were able to BLAST these against NCBI and get hits against the NCBI public sequence database and get Silene hits straight back. We were also able to map them onto the Silene genome directly using BWA, even with no trimming or masking low-quality regions. Overall, we got nearly a full week’s sequencing from our flowcell, with ~24,000 reads at a mean length of ~4kb. Not bad, and we’d have got more if we hadn’t wasted the best part of the run sodding about while we troubleshot our library mix-up at the start (the number of active pores, and hence sequencing throughput, declines over time).
The bad: There is a price for these long, rapid reads. Firstly, the accuracy is definitely lower than Illumina – although the average lambda library sequencing accuracy in our first library was nearly 90%, on some of our mapped Silene reads it dropped much lower – 70% or so. This has been well documented, of course. Secondly, the *big* difference between the MinION in use and a MiSeq or HiSeq is the sheer level of involvement needed to get the best from the device – whereas both Illumina machines are essentially push-button operated, the MinION seems to respond well if you treat it kindly (reloading etc) and not if you don’t (introducing air bubbles while loading seemed to wreck some pores)
The very, very, good: Ultimately though, we came away convinced that these issues won’t matter at all in the long run. ONT have said that the library prep and accuracy are both boing improved, as are flowcell quality (prototype, remember). Most importantly the MinION isn’t really a direct competitor to the HiSeq. It’s a completely different instrument. Yes, they both read DNA sequences, but that’s where the similarity ends. The MinION is just so flexible, there’s almost an infinite variety of uses.
In particular, we were really struck that the length of the reads means simple algorithms like BLAST can get a really good match from any sample with just a few hundreds, or tens, of reads from a sample. You just don’t need millions of 150bp reads to match an unknown sample to a database with reads this long! Coupled with the fact that the software is real-time and flowcells can be stopped and started, and you have the bones of a really capable genetic identification system for all sorts of uses; disease outbreaks (in fact see this great work on Ebola using MinION); customs control of endangered species; agriculture; brewing – virtually anything, in fact, where finding out about living organisms’ identity or function is needed.
The future.
There’s absolutely no question at all: really, these devices are the future of biology. Maybe the MinION will take off (right now, the buzz couldn’t be hotter), or a competitor will find a way to do even more amazing things. It doesn’t matter how it comes about, though – the next generation of biologists really will be living in a world where, in 10, 15, 20 years, at most, the sheer ubiquity of sequencers like this will mean that most, if not all eukaryotes’ genomes will have been sequenced, and so can be easily matched against an unknown sample. If that sounds fantastic, consider that the cumulative sequencing output of the entire world in 1988 amounted to a little over 100,000 sequences, a yield equivalent to a single good MinION run. And while most people in 1988 thought that, since the human genome might take 30 or 40 years to sequence, there was little point in even starting, a few others looked around at the new technology, its potential, and drew a different conclusion. The rest is history…